Greatest Hits
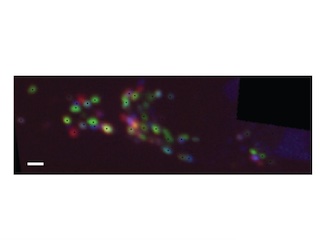
Fast deep neural correspondence for tracking and identifying neurons in C. elegans using semi-synthetic training
With Xinwei Yu and the Leifer Lab. eLife, 2021
We present an automated method to track and identify neurons in C. elegans using Transformer networks.
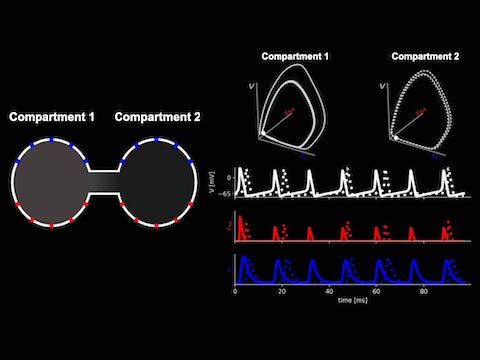
Scalable Bayesian inference of dendritic voltage via spatiotemporal recurrent state space models
With Ruoxi Sun, Ian Kinsella, and Liam Paninski.
NeurIPS, 2019
Oral Presentation
Recurrent SLDS models for smoothing voltage imaging data.
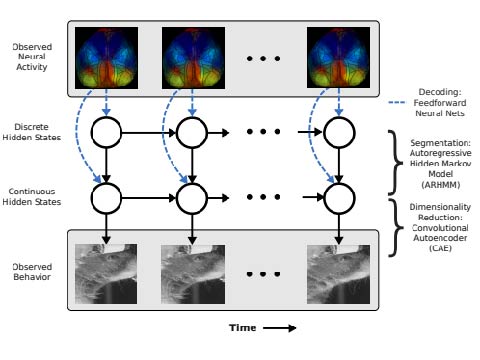
BehaveNet: nonlinear embedding and Bayesian neural decoding of behavioral videos
With Ella Batty, Matt Whiteway, Liam Paninski, and many others. NeurIPS, 2019
Combining convoluational autoencoders and autoregressive hidden Markov models for neural and behavioral data.
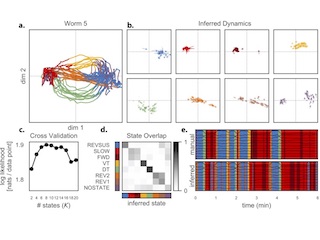
Hierarchical Recurrent State Space Models of Neural Activity
With Annika Nichols, David Blei, Manuel Zimmer, and Liam Paninski. bioRxiv, 2019
We develop hierarchical and recurrent state space models for whole brain recordings of neural activity in C. elegans. We find states of brain activity that correspond to discrete elements of worm behavior and dynamics that are modulated by brain state and sensory input.
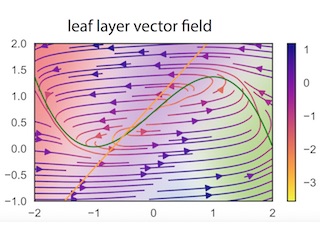
Tree-structured Recurrent SLDS
With Josue Nassar, Monica Bugallo, and Il Memming Park. ICLR, 2019
We develop an extension of the rSLDS to capture hierarchical, multi-scale structure in dynamics via a tree-structured stick-breaking model. We recursively partition the latent space to obtain a piecewise linear approximation of nonlinear dynamics. A hierarchical prior smooths dynamics estimates, and inference is performed via an augmented Gibbs sampling algorithm.
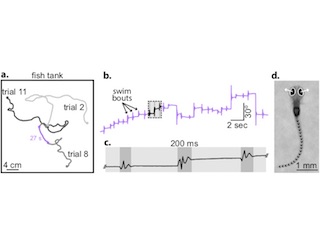
Point process latent variable models of larval zebrafish behavior
With Anuj Sharma, Robert Johnson, and Florian Engert. NeurIPS 2018
We develop deep state space models with point process observation models to capture structure in larval zebrafish behavior. The models combine discrete and continuous latent variables. We marginalize the discrete states with message passing and perform inference with bidirectional LSTM recognition networks.
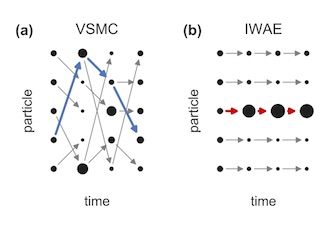
Variational Sequential Monte Carlo
With Christian Naesseth, Rajesh Ranganath, and David Blei. AISTATS 2018
We view SMC as a variational family indexed by the parameters of its proposal distribution and show how this generalizes the importance weighted autoencoder. As the number of particles goes to infinity, the variational approximation approaches the true posterior.
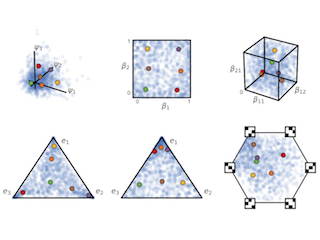
Reparameterizing the Birkhoff Polytope for Variational Permutation Inference
With Gonzalo Mena, Hal Cooper, Liam Paninski, and John Cunningham. AISTATS 2018
How to perform gradient-based variational inference over permutations and matchings via a continuous relaxation to the Birkhoff polytope.
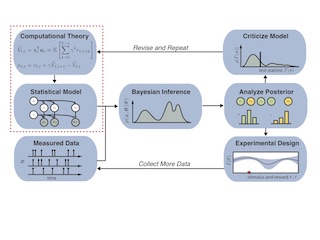
Using Computational Theory to Constrain Statistical Models of Neural Data
With Sam Gershman. Current Opinion in Neurobiology, 2017
Top-down and bottom-up methods are joined in a theory-driven analysis pipeline. We view theories as priors for statistical models, perform Bayesian inference, criticize, and revise.
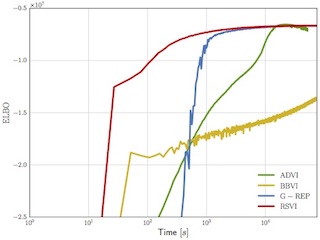
Rejection Sampling Variational Inference
With Christian Naesseth, Fran Ruiz, and David Blei.
AISTATS 2017
Best Paper Award
Reparameterization gradients through rejection samplers for automatic variational inference in models with gamma, beta, and Dirichlet latent variables.
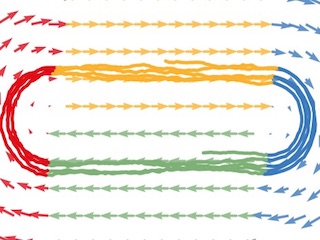
Recurrent Switching Linear Dynamical Systems
With Matt Johnson, Andy Miller, Ryan Adams, David Blei, and Liam Paninski. AISTATS, 2017
Bayesian learning and inference for models with co-evolving discrete and continuous latent states.
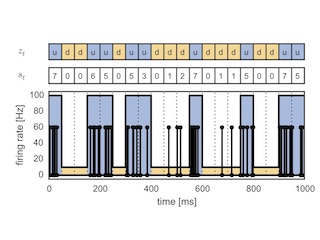
Bayesian Methods for Discovering Structure in Neural Spike Trains
2016 Leonard J. Savage Award
My dissertation work at Harvard University on networks, point processes, and state space models for neural data analysis.
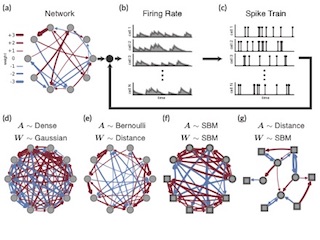
Uncovering Structure in Neural Data with Networks and GLMs
With Ryan Adams and Jonathan Pillow. NIPS 2016
We combine network priors, nonlinear autoregressive models, and Pólya-gamma augmentation to reveal latent types and features of neurons using spike trains alone.
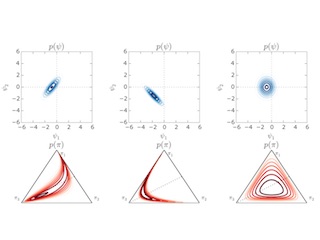
Dependent Multinomial Models Made Easy
With Matt Johnson and Ryan Adams. NIPS 2015
We use a stick-breaking construction and Pólya-gamma augmentation to derive block Gibbs samplers for linear Gaussian models with multinomial observations.
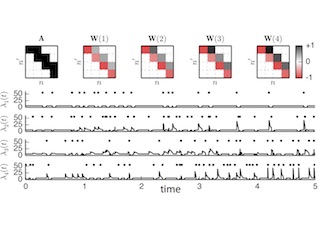
Studying Synaptic Plasticity with Time-Varying GLMs
With Chris Stock and Ryan Adams. NIPS 2014
We propose a time-varying generalized linear model whose weights evolve according to synaptic plasticity rules, and we perform Bayesian inference with particle MCMC.
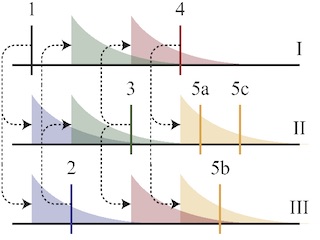
Discovering Latent Network Structure in Point Process Data
With Ryan Adams. ICML 2014
Combining Hawkes processes with generative network models to uncover latent patterns of influence.